By. Natalie Reyna
A look at microbes in the environment through the latest scientific research. - The Environmental Microbiology students, TAMIU
Monday, November 27, 2023
Environmental adaptation of host and a microbiome
Wednesday, November 1, 2023
Overcoming contamination in hospital: Finding hidden source of bacteria in a radiopharmacy.
By: Albert Gonzalez
Image of the locations sampled for bacterial content on surfaces in the hospital's radiopharmacy. Each red diamond shows the sampled location, and the arrows display the bacterial contamination's movement.Hospitals need to be as clean as possible; the patients' health rests on it. This being said, preventing bacterial contamination in a hospital environment is essential. A recent investigation surrounding sudden and persistent bacterial contamination of a hospital's radiopharmacy was conducted. Radiopharmacies are the section of the hospital that holds radioactive medicine. These medicines are highly important; protecting them from contamination is vital to their maintenance. In this investigation, the radiopharmacy unit of a university teaching hospital searched for the source of an ongoing bacterial contamination. They did this by testing the bacterial content of many surfaces. An interesting new method they used involved placing sterile water into a container, shaking it, and testing it to see what kinds of bacteria were on the surfaces of the container. The unit discovered that the contamination source came from reusable plastic containers. From the data gathered through this investigation, the unit realized the potential of old plastic containers to be a source of bacterial contamination and the effectiveness of a method for sampling surfaces for bacteria. These findings can be shared with other hospitals and help keep them free from contamination, ultimately ensuring patients' health.
Original article: Armando, M., Barthelemi, L., Couret, I., Verdier, C., Dupont, C., Jumas-Bilak, E., Grau, D., (2022) Recurrent environmental contamination in a centralized radiopharmacy unit by Achromobacter spp: Results of a large microbiological investigation. American Journal of Infection Control, 51, 5., 557-562.
Ions Effectively Inhibit Airborne Bacteria Viability
By: Destinee Lopez
Figure 1 (A and B) S. aureus (A) and E. coli (B) plated at 104 CFU/ml on 150-mm petri dishes were subjected to a direct ion effect, with the ionizer positioned 5 or 10 cm away. n ≥ 3 duplicate studies. *, Student's t-tests; P < 0.05.Human health depends on indoor air quality, and since airborne pathogens can be harmful, there is growing interest in air ionization technology as a potential means of reducing bacterial growth and improving overall air quality. This study investigates the impact of ion exposure on the survival of Staphylococcus aureus and Escherichia coli bacteria, regardless of whether the bacteria is trapped in air filters or plated on agar. Potential variations in the effects of exposure are examined, which may be influenced by factors such as filter type, action area, distance from the ion generator, bacterial type and load, and exposure time. This study supports the potential use of air ionizers for preventing and controlling indoor airborne infections by offering essential insights into the solid antibacterial activity of air ions and demonstrating their effectiveness in lowering the viability of common airborne pathogens, such as S. aureus and E. coli, under a variety of conditions. The purpose of the experiment was to determine how different experimental conditions, such as bacterial type and load, action area, distance from the ion generator, exposure time, and filter type, affected the viability of Staphylococcus aureus and Escherichia coli bacteria, both plated on agar and trapped in air filters. The results supported the potential use of air ionizers for preventing and controlling indoor airborne infections by showing a significant and consistent antibacterial activity of both positive and negative ions, which reduced the viability of both Staphylococcus aureus and Escherichia coli bacteria, whether plated on agar or trapped in air filters, across various experimental conditions.
Original article: Comini, Mandras, N., Iannantuoni, M. R., Menotti, F., Musumeci, A. G., Piersigilli, G., Allizond, V., Banche, G., & Cuffini, A. M. (2021). Positive and Negative Ions Potently Inhibit the Viability of Airborne Gram-Positive and Gram-Negative Bacteria. Microbiology Spectrum, 9(3), e0065121–e0065121. https://doi.org/10.1128/Spectrum.00651-21
Dustborne vs Airborne Fungi in Domestic Environments
By: Esmeralda Sandoval
In recent years, poor aeration is a major cause of increasing microbial concentrations in domestic environments because of limited air exchange and removal of indoor pollutants due to windows not being left open in efforts to maintain comfortable climate conditions indoors. Dust serves as reservoir for fungi, allowing it to proliferate in high humidity conditions by using its organic matter. The dustborne fungi can then be suspended into the air and become airborne. In this study conducted by Pyrri and colleagues, fungal concentration were examined from the air and dust during the winter and summer months to determine the negative implications posed to human health. The results showed that fungal concentrations were higher in the summer, averaging at 931 CFU m-3 in the air, than in the winter, averaging at 739 CFU m-3. These concentrations are higher than the suggested level by the World Health Organization (WHO), 150 CFU m-3. These results indicate that when dust is not frequently removed, fungal concentrations increase allergen and pathogen exposure to humans.
Original Article:
Pyrri, I., Stamatelopoulou, A., Pardali, D., & Maggos, T. (2023). The air and Dust Invisible mycobiome of urban domestic environments. Science of The Total Environment, 904, 166228. https://doi.org/10.1016/j.scitotenv.2023.166228
Double Trouble Oil Degradation
By: Raul Gonzalez
Harmful algal blooms
Algal Blooms are a naturally occurring phenomena in freshwater and marine environments, blooms can be an essential part of an ecosystem, providing food and oxygen for the environment. However, human intervention has allowed algal blooms to become an environmental hazard. In the last 30 years, Factors such as climate change, agricultural runoff, and sewage pollution, have led to the harmful algal blooms to increase in frequency and intensity, leading to the formation of “Dead Zones”. Dead Zones are named as such due to their low oxygen content and have caused species of fish to die off and negatively impacting the food web by causing the small organisms such as plankton to die off as well. The ability of certain species of algae to produce toxins can also negatively affect humans who are exposed to the infected bodies of water, not to mention the risk it poses to pets and livestock. Despite the fact the impact of blooms is detrimental to most environments they occur in, little is being done in the way of remedying the issue.
Influence of the Rama Stable Isotope approach
Original Article
Anal. Chem.2020, 92, 16, 11429–11437 Publication Date: July 22, 2020 https://doi.org/10.1021/acs.analchem.0c02443
Reprint of Organic waste conversion through anaerobic digestion: A critical insight into the metabolic pathways and microbial interactions

Figure 1. Description of how waste becomes analytical platforms, anaerobic bioprocess turned to two different processes, and valuables becoming analytical data
Anaerobic digestion is the process where organic waste is broken down into biogas since it is the best way where no harm is done to the environment. Anaerobic digestion is a process where it includes four biochemical phases such as: hydrolysis, acidogenesis, acetogenesis, and methanogenesis. Hydrolysis is the process where the greater molecules are transformed to smaller ones. Acidogenesis is the process where hydrolytic products such as carbon, hydrogen, and oxygen are then turned into volatile fatty acids such as butyric acids. Acetogensis is the process where its products are converted into hydrogen as well as acetate. In this study, the methods used include an explanation of how the pathways are arbitrated from bacteria and archaea are important due to the relationship of the communities. The results that were concluded by the scientists explain that the chemical conversion such as the conversion of microbes to methane, or the conversion to acetic acid were important for the discoveries in regards to anaerobic digestion. In conclusion, anaerobic digestion is known what it is today due to the studies of the four biochemical pathways and the microbes. The authors concluded that in order to come to a final discussion, hydrogenotrophic using 30% of methane and acetoclastic uses 70% of methane which are necessary for anaerobic digestion.
References
Monika Yadav, Chandrakant Joshi, Kunwar Paritosh, Jyotika Thakur, Nidhi Pareek, Shyam Kumar Masakapalli, Vivekanand Vivekanand, Reprint of Organic waste conversion through anaerobic digestion: A critical insight into the metabolic pathways and microbial interactions, Metabolic Engineering, Volume 71, 2022, Pages 62-76, https://www.sciencedirect.com/science/article/abs/pii/S1096717622000258
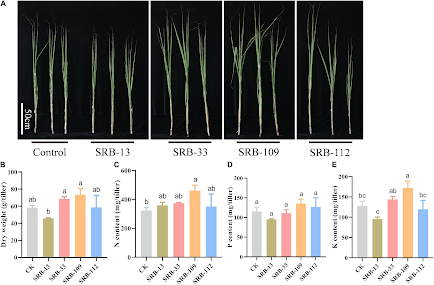
From Soil to Sweetness: The Role of Bacteria in Sugarcane's Thriving Journey
By: Kenya Luquin
Leprosy-causing bacterium's DNA detected in soil
Leprosy is a contagious illness caused by the Mycobacterium leprae organism. This disease affects the skin and nerves of infected humans, which is the reason why it is considered a major health threat. In a study conducted by Tió-Coma and colleagues (2019), it was investigated whether the DNA of this organism could be found in the soil of regions where leprosy is common. To detect the presence of this organism in the collected samples, polymerase chain reaction (PCR) amplification was performed. PCR is a highly specific laboratory technique utilized to make copies of a DNA segment. This allows scientists to identify genetic material by detecting and amplifying a region of the bacterium's genetic sequence. The results revealed that the DNA of M. leprae was present in the soil in proximity to houses of leprosy patients in Bangladesh. It was also discovered in the holes of armadillos located in Suriname, and in the habitat of leprosy-carrying red squirrels in the British Isles. This suggests that the DNA of M. leprae can be found in the soil, indicating that the environment could serve as a temporary bank for this bacteria.
Original article: Tió-Coma, M., Wijnands, T., Pierneef, L. et al. Detection of Mycobacterium leprae DNA in soil: multiple needles in the haystack. Sci Rep 9, 3165 (2019). https://doi.org/10.1038/s41598-019-39746-6
Nectar Inhabiting Bacteria's Affect on Olfactory Responses of Insect Parasitoids by Altering Nectar Odors
By Victoria Barrera 
The figure demonstrates the Olfactory response of adult T. basalis females when given the choice between test and control odors. Test odors consisted of either buckwheat raw or synthetic nectar.
Produced by flowering plants, floral nectar is a sugar-rich resource colonized by various microorganisms that alter physical and chemical traits including scent due to production of microbial volatile organic compounds (mVOCs). Nectar is mainly studied for its role in attracting insects such as parasitoids, commonly found visiting and feeding on nectar of various plants. That being said, mVOC’s effect on parasitoid's olfactory responses to flowering nectar is often overlooked. Currently, researchers aim to record if bacteria from phyla: Firmicutes (8 isolates), Proteobacteria (4 isolates), and Actinobacteria (2 isolates) associated with the nectar of Fagopyrum esculentum (Buckwheat) affect the response of Trissolcus basalis (wasp species) via odor changes. Nectar from 50 F. esculentum flowers were collected and 100 µL from 20 samples were placed onto a trypticase soy agar (TSA) for isolation. T. Basalis females were used in all bioassays and were individually placed in small vials 24 hours prior to induce starvation. Their results found that 4 out of 14 strains attracted T. Basalis, indicating that nectar-inhabiting bacteria affect interactions between flowering plants and parasitoids. Correspondingly, these results are relevant to biological control of insect pests as nectar modifications may enhance the future progression of human agriculture/pesticides.
Original Article:
Cusumano A., Bella P., Peri E. et al. Nectar-Inhabiting Bacteria Affect Olfactory Responses of an Insect Parasitoid by Altering Nectar Odors. Microb Ecol 86, 364–376 (2023).
What we leave behind; The alternative and stabilizing treatment for abandoned waste.
By. Carolina Perez
The image shows both abandoned mine tailings sites, an analysis of anaerobic sludge acclimation for sulfate reduction, and the set up for biological treatment of these mine tailings. Figure taken from E. I. Valenzuela et al. 2020.
The mining industry can provide many metal compounds used
for financing, technology, and many more consumer products. Although, mining
underground can cause waste of rocks, residue from the overall mining
production and tailings. This type of waste can lead to potential hazards based
on the environmental atmosphere, the number of deadly particles exposed, and
the timing for which the mine was created. Tailings are most concerning as they
contain a high concentration of metals and soluble salts. The alternative processes of sulfate reducing factors were examined as a
possible treatment for stabilizing hazardous metal elements. This study
included two samples from two different abandoned mine tailings of known
activity within Sonora, Mexico. With several treatments the mine tailings were
utilized as a solid phase that supplies sulfate in support of sulfate reducing
activities, which stabilizes harmful components by the precipitation of
metallic sulfide. The end results initiated the toxic
metals concerning the environment can relocate from an aqueous phase to a
residual phase. Restricting the release of these metals in natural solution and
depleting their impact on the environment. It was concluded that sulfate
reducing processes can be an effective stabilizer toward collateral pollution
caused by residue waste in abandoned mines. If these exposed metals were to be
left abandoned and unchecked without treatment, the most at risk will be
environmental ecosystems and human health.
Original
article
Valenzuela, E. I.,
García-Figueroa, A. C., Amábilis-Sosa, L. E., Molina-Freaner, F. E., &
Pat-Espadas, A. M. (2020). Stabilization of potentially toxic elements
contained in mine waste: A microbiological approach for the environmental
management of mine tailings. Journal of Environmental Management, 270,
110873. https://doi.org/10.1016/j.jenvman.2020.110873
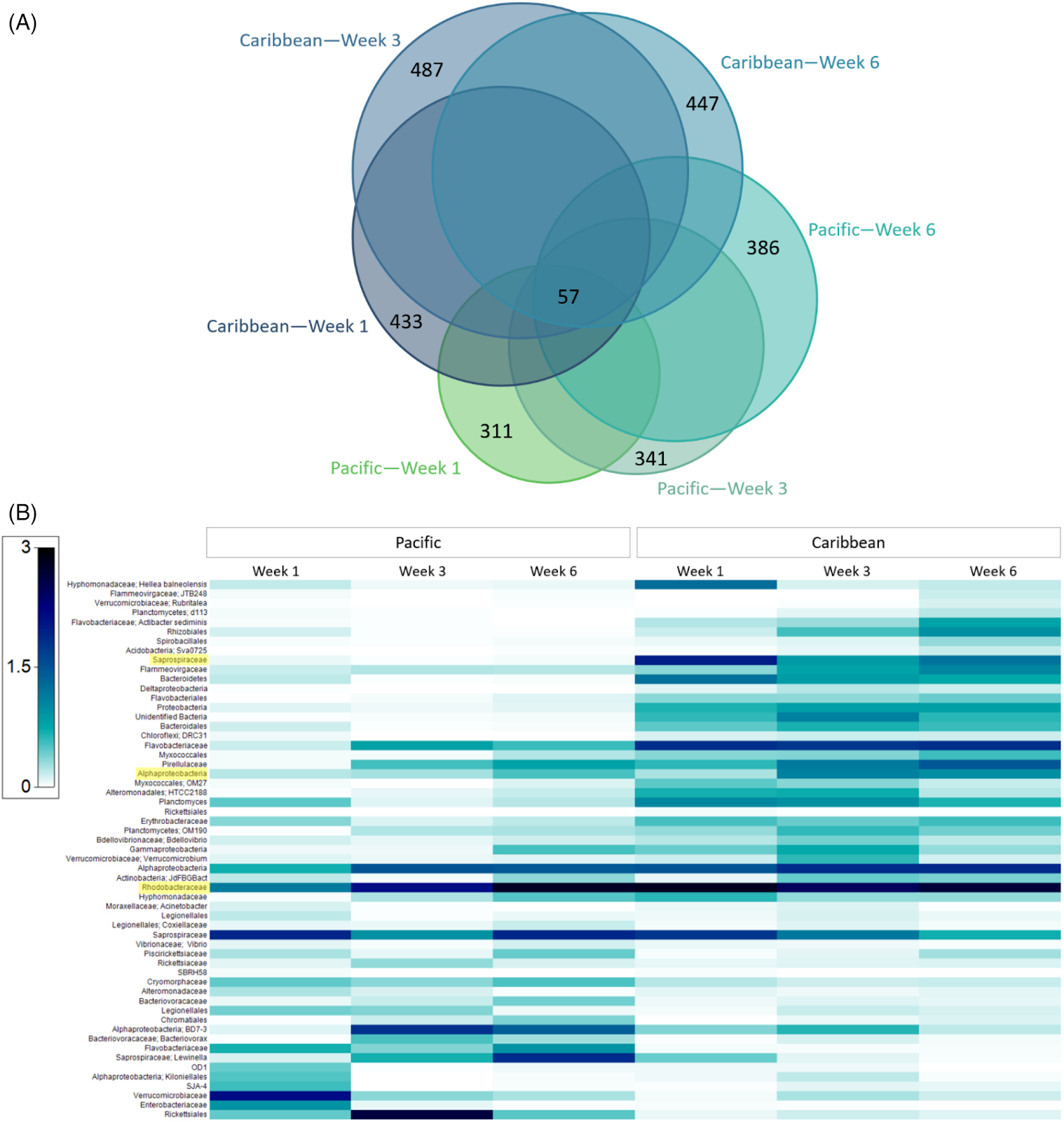
Environmental exposure to plastics shapes the marine microplastic - associated bacterial communities.
Unraveling the Mysteries of Cyanobacterial Toxins: Implications for Water Safety
In the research article “Cyanobacterial toxins in water,” the enigmatic world of cyanobacterial toxins is unveiled, shedding light on their significance in water safety. These toxins, produced by cyanobacteria (blue-green algae), have long been known to be harmful to animals, yet their impact on human health remained elusive until recently. This study reveals that toxic cyanobacteria are more prevalent in water bodies than previously believed, raising concerns about drinking water safety worldwide. The research highlights the diverse range of symptoms in animals and humans, emphasizing the urgency for increased awareness and monitoring. The scientists also explored factors influencing toxin formation, offering crucial insights into the regulation of these harmful compounds. This study is a vital step toward safeguarding public health and necessitates further research to develop strategies for toxin detection and removal, ensuring the purity of our water sources. As society grapples with water scarcity and increasing demand, this research holds paramount importance for policymakers, environmental scientists, and health professionals, guiding future practices to preserve our most precious resource, clean water.
Original article:
Codd, G. A., Bell, S. G., & Brooks, W. P. (1989). Cyanobacterial toxins in water. Water Science and Technology, 21(3), 1–13. https://doi.org/10.2166/wst.1989.0071
Cyanobacteria Blooms and the reaction with Dynamic Microbiomes
In nature, the natural process of bacteria persisting in fresh water environments depends on the effect of weather, organisms, etc. In the laboratory, scientist are able to manipulate the reduction of contaminants and creating pure cultures. Without microbiomes and the relationship of cyanobacteria the process is limited and complications may occur in research. Microcystis aeruginosa NIES-843 cultures were in light limited control with different sources and reduced conditions in samples that were cultivated identical microbiomes for 3 cycles of growth. On the right hand side on the image, we are able to depict that there is nutrient availability from the shift in microbiomes. Specifically, Microcystis aeurginosa NIES-843 cultures that were started from the same stock and transferred for 3 generations to Nitrogen and Phosphorus conditons to experiment how cyanobacterium would react (Steffen et al. 2014). A series of questions such as to what extent can a microbe's microbiome affect gene expression & what biological pattern can scientists expect to see in nature in combination with cell interaction, chemistry, physics, etc. within changing environments.
Pound, Helena L., et al. “Environmental Studies of Cyanobacterial Harmful Algal Blooms Should Include Interactions with the Dynamic Microbiome.” Environmental Science & Technology, 16 Sept. 2021, https://doi.org/10.1021/acs.est.1c04207.
Oil Spill Clean Up
By: Jessica Rodriguez
Oil spills can harm shorelines, animals, ocean water and floors. Oil is toxic to fish embryos and their developmental stages are sensitive to oil exposure and about 100% of them die when exposed to it. The reduction of fish population has an impact on the food source for people and all marine life. SQT (Sediment quality triad), are three ways to determine treatment for animals that live in ocean waters and floors that are at risk of harmful chemicals, potential toxic affects to rocks and sediments that are sensitive to fuel chemicals, and health of all marine life. There are biological and physical treatments to help clean up oil spills. Physical treatment of hot water and high-pressure flushing resulted in the maximum removal of oils by (93%). Fertilizer, chemical agents, stimulants, and bacterial growth are examples of biological treatments that improved environment recovery and removal of oil by (66%). It has been found there are bacteria such as Dietzia sp. and Rosevarius sp. that grow in marine environments that can help break down oil residues from oil spills. Together, biological treatments and promoting the growth of these bacteria have helped to reduce residual oil. More research is required to decide between biological and physical remediation, as well as whether applying bacteria capable of breakdown of oil is required for an oil spill response.
The figure shows SQT and physical & chemical treatment techniques in oil spills. The figure is from Kim et al. (2022)